Command Line Interface
Contents
Command Line Interface¶
metaDMG has the following commands:
Config¶
metaDMG config takes a single argument, samples, and a bunch of additional options and flags.
The samples refer to a single or multiple alignment-files (or a directory containing them), all with the file extensions: .bam, .sam, and .sam.gz.
Parameters¶
Damage mode¶
--damage-mode:[lca|local|global].lcais the recommended and automatic setting. If usinglocal, it means that damage patterns will be calculated for each chr/scaffold contig. If usingglobal, it means one global estimate. Note that when using[local|global]the LCA parameters won’t matter.
LCA¶
Options:
--names: Path to the (NCBI)names-mdmg.dmp. Mandatory for LCA.--nodes: Path to the (NCBI)nodes-mdmg.dmp. Mandator for LCA.--acc2tax: Path to the (NCBI)acc2tax.gz. Mandatory for LCA.--min-similarity-score: Normalised edit distance (read to reference similarity) minimum. Number between 0-1. Default: 0.95.--max-similarity-score: Normalised edit distance (read to reference similarity) maximum. Number between 0-1 Default: 1.0.--min-edit-dist: Minimum edit distance (read to reference similarity). Positive integer. Note that edit distances scores cannot be set at the same time as similarity scores; choose one or the other.--max-edit-dist: Maximum edit distance (read to reference similarity). Positive integer. Note that edit distances scores cannot be set at the same time as similarity scores; choose one or the other.--min-mapping-quality: Minimum mapping quality. Default: 0.--lca-rank: The LCA rank used in ngsLCA. Can be eitherfamily,genus,speciesor""(everything). Default is"".
Flags:
--custom-database: Using a custom database or the NCBI. If NCBI, automatically corrects for a couple of bad taxa. Default is False.
General¶
Options:
--output-dir: Path where the generated output files and folders are stored. Default:./data/.--config-file: The name of the generated config file. Default:config.yaml.--metaDMG-cpp: The command needed to run themetaDMG-cppprogram.--max-position: Maximum position in the sequence to include. Default is (+/-) 15 (forward/reverse).--min-reads: Minimum number of reads to include in the fits (min_reads <= N_reads)..--parallel-samples: The number of samples to run in parallel. Default is running in seriel.--cores-per-sample: Number of cores to use pr. sample. Do not change unless you know what you are doing.--sample-prefix: Prefix for the sample names.--sample-suffix: Suffix for the sample names.--weight-type: Method for calculating weights. Default is 1. Do not change unless you know what you are doing.
Flags:
--forward-only: Only fit the forward strand.--bayesian: Include a fully Bayesian model (probably better, but also a lot slower, about a factor of 100).--long-name: Use the full, long, name for the sample.--overwrite: Overwrite config file without user confirmation.
Examples¶
$ metaDMG config raw_data/alignment.sorted.bam \
--names raw_data/names-mdmg.dmp \
--nodes raw_data/nodes-mdmg.dmp \
--acc2tax raw_data/acc2taxid.map.gz \
--parallel-samples 4
metaDMG is pretty versatile regarding its input argument and also accepts multiple alignment files:
$ metaDMG config raw_data/*.bam [...]
or even an entire directory containing alignment files (.bam, .sam, and .sam.gz):
$ metaDMG config raw_data/ [...]
To run metaDMG in non-LCA mode, an example could be:
$ metaDMG config raw_data/alignment.sorted.bam --damage-mode local --max-position 15 --bayesian
Config GUI¶
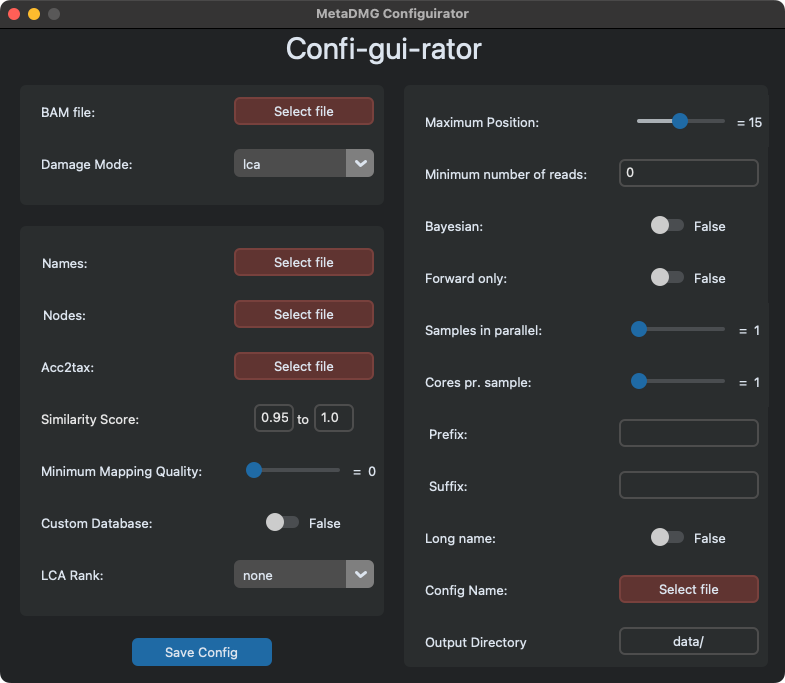
metaDMG config-gui is a simple graphical user interface (GUI) to help with the config creation.
The command itself does not take any parameters, everything is done by clicking and dragging.
For more information about what the different buttons and sliders mean, see the normal config command.
Examples¶
$ metaDMG config-gui
The GUI presented looks like this:
Mandatory fields that need to be filled are coloured red.
Note that if you change the damage mode to LOCAL or GLOBAL, the bottom left
square becomes disabled, since these parameters are only relevant for LCA.
Compute¶
The metaDMG compute command takes an optional config-file as argument
(defaults to config.yaml if not specified).
Parameters¶
Flags:
--force: Forced computation (even though the files already exists).
Dashboard¶
You can now see a preview of the interactive dashboard.
The metaDMG dashboard command takes first an optional config-file as argument
(defaults to config.yaml if not specified).
Parameters¶
Options:
--results: Path to the results directory.--port: The port to be used for the dashboard. Default is8050.--host: The dashboard host adress. Default is0.0.0.0.
Flags:
--debug: Allow for easier debugging the dashboard. For internal usage.--server: If running on a server
Examples¶
$ metaDMG dashboard
$ metaDMG dashboard non-default-config.yaml --port 8050 --host 0.0.0.0
Get Data¶
The metaDMG get-data command gets test data and saves it in the output-dir. Useful for e.g. the online tutorial.
Parameters¶
Options:
--output-dir: Path to the output directory.
Convert¶
The metaDMG convert command takes first an optional config-file as argument
(defaults to config.yaml if not specified) used to infer the results directory.
Parameters¶
Options:
--results: Direct path to the results directory.--output: Mandatory output path.
Flags:
--add-fit-predictions: Include fit predictions D(x) in the output.
Note that neither the config-file nor --results have to be specified
(in which just the default config.yaml is used), however,
both cannot be set at the same time.
Examples¶
$ metaDMG convert --output ./directory/to/contain/results.csv
$ metaDMG convert non-default-config.yaml --output ./directory/to/contain/results.csv --add-fit-predictions
Filter¶
The metaDMG filter command takes first an optional config-file as argument
(defaults to config.yaml if not specified) used to infer the results directory.
Parameters¶
Options:
--results: Direct path to the results directory.--output: Mandatory output path.--query: The query string to use for filtering. Follows the Pandas Query() syntax. Default is""which applies no filtering and is thus similar to themetaDMG convertcommand.
Flags:
--add-fit-predictions: Include fit predictions D(x) in the output.
Note that neither the config-file nor --results have to be specified
(in which just the default config.yaml is used), however,
both cannot be set at the same time.
Examples¶
$ metaDMG filter --output convert-no-query.csv # similar to metaDMG convert
$ metaDMG filter --output convert-test.csv --query "N_reads > 5_000 & sample in ['subs', 'SPL_195_9299'] & tax_name == 'root'" --add-fit-predictions
Plot¶
The metaDMG plot command takes first an optional config-file as argument
(defaults to config.yaml if not specified).
Parameters¶
Options:
--results: Direct path to the results directory.--query: The query string to use for filtering. Follows the Pandas Query() syntax. Default is""which applies no filtering.--samples: A comma-space separated string containing the samples to use in the plots. Default is""which applies no filtering.--tax-ids: A comma-space separated string containing the tax-ids to use in the plots. Default is""which applies no filtering.--output: The path to the output pdf-file. Defaults topdf_export.pdf.
Examples¶
$ metaDMG plot
$ metaDMG plot --query "100_000 <= N_reads & 8_000 <= phi" --tax-ids "1, 2, 42" --samples "sampleA, another-sample" --pdf-out name-of-plots.pdf
PMD¶
The metaDMG PMD command takes an alingment file as argument and computes the PMD scores for each read in the file. The results are saved to a csv file.
Examples¶
$ metaDMG PMD raw_data/alignment.sorted.bam --output PMDs.csv --metaDMG-cpp ./metaDMG-cpp
mismatch-to-mapDamage¶
The metaDMG mismatch-to-mapDamage command takes a mandatory mismatch-file as argument
and converts it to the mapDamage format misincorporation.txt.
Parameters¶
Options:
--csv-out: Output CSV file (misincorporation.txt). Default ismisincorporation.txt.
Examples¶
$ metaDMG mismatch-to-mapDamage data/mismatches/XXX.mismatches.parquet
$ metaDMG mismatch-to-mapDamage data/mismatches/XXX.mismatches.parquet --csv-out misincorporation.txt